Audits are probably the most dreaded aspect of NGS data analysis, and like specters, they come in many forms to haunt you.
You might need to publish results from several years back, only to realize that the open source tools you used have dramatically changed, forcing you to find version numbers and any older features that might have not followed updated best practices. There goes any reproducibility.
You might be required to prepare for an FDA audit, or asked to follow CDC guidelines. That means following the breadcrumbs across your NGS data analysis stack, ensuring everything is compliant.
Or, your lab has hired a temp bioinformatician, and a year down the line you have no idea what tools they used!
These are just a few of many such scenarios we’ve heard from our peers over the years. Our founder, Amit, was an instructor at Harvard Medical School prior to starting Basepair. With over 40 published papers, he gets that transparency, traceability, and QC are not just convenient – they save researchers, bioinformaticians – and really, entire labs – from future headaches.
In this post, we’ll overview how our team audits themselves, and how this translates to a more transparent, audit-ready platform with reproducible results.
How Basepair ensures its platform is audit-proof
From aligning reads for DNA-Seq data, to running DESeq on RNA-Seq results, to performing motif analysis on a ChIP-Seq dataset, each algorithm in our custom workflows is rigorously tested on public datasets.
We are continuously reviewing the latest research reports that compare new methods in NGS data analysis, while ensuring our tools are in line with regulations like ACMG’s clinical laboratory standards.
(We publish any interesting findings from our reviews on our blog – we recommend you check out Performing pathway enrichment with non-human species, and Trimming for RNA-Seq data.)
Staying up-to-date with the latest data analysis tools and techniques is a difficult task for a bioinformatician, and for biologists it’s pretty much out of scope. As a team of bioinformatics PhDs and developers, this is our full-time job.
Basepair’s pipeline architecture automatically stores every software version, reference file, and parameter value, and other useful metadata. These data are available as a log file, providing full providence of data flow.
We have a feature that allows you to download a log of all the tools and methods that were used in your workflow. Try it out the next time you run an analysis!
Let’s see how this translates to a smoother QC process, and more transparency and reproducibility for you and your team.
QC and transparency in Basepair
Alongside rigorous assessment of the latest tools and techniques, and adherence to the latest best practices and guidelines, we feature several tools we have found to be helpful for ensuring your results are ready for publication or further downstream analysis.
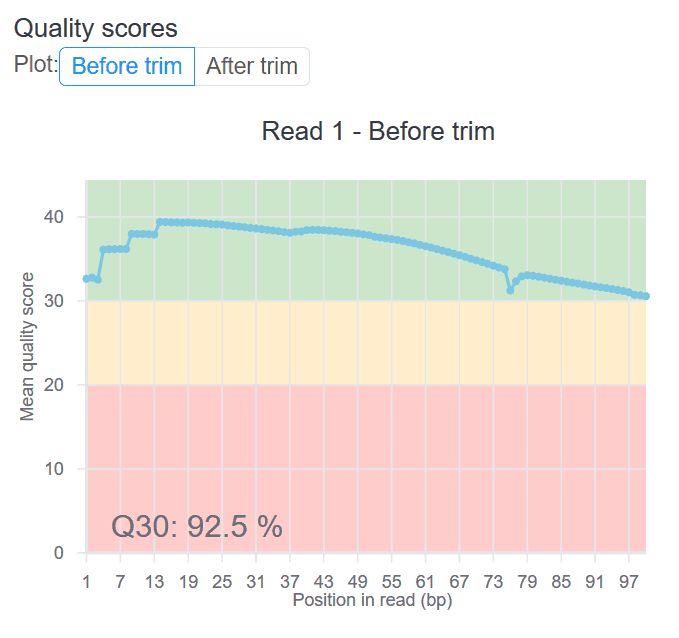
QC scores are available in most of our workflows, and are tailored to each. For example, here is a screenshot of the before/after trim QC chart for the STAR-based expression count workflow on RNA-Seq data:
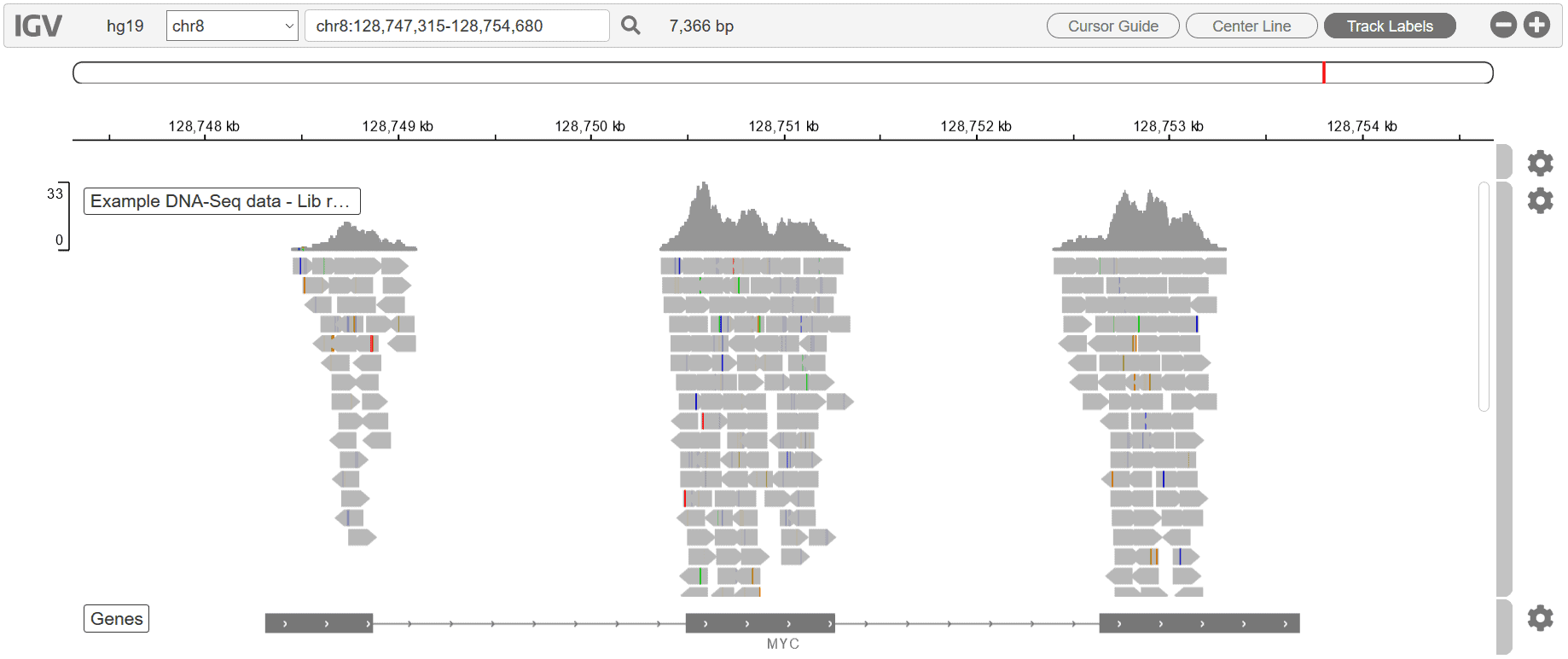
 Besides these downloadable, publication-ready graphs, we also include a Genome Browser in every report, for every workflow:
Besides these downloadable, publication-ready graphs, we also include a Genome Browser in every report, for every workflow:
The Genome Browser allows for an overarching visual review of every gene in your raw dataset. To learn more about our QC charts, Genome Browser, and various other report features, see our blog, Visualizing your NGS results with Basepair.
You can try these features yourself with a free Basepair trial – we include several sample reports where you can inspect QC charts and interact with the Genome Browser. Or, you can upload your own data to analyze!