Want to learn more about variant calling? Take a look at our most recent webinar: Variant Calling for Bench Scientists: Tools, Tips & Best Practices
Automated DNA sequence analysis for researchers
Zero code knowledge required
Never used NGS data analysis tools? No problem. Basepair’s automated NGS data analysis platform requires zero lines of code to run DNA sequence analysis, no matter how complicated the workflow.
Publication-ready visual reports
High-quality interactive figures and flexible data-exploration tools are available upon analysis completition, so you can get to downstream analysis – or publication – faster. No fine-tuning or file manipulation necessary.
Unlimited scalability
Planning to run a single sample or a thousand? No matter how many DNA-Seq samples you need to analyze, Basepair is able to automatically scale to your workload. Finish analyses in hours, not days or weeks.
What if you could finish your DNA-Seq data analysis with a single click?
Basepair’s whole genome/exome analysis pipelines take minutes to set up, and hours (or less) to run. Reports are auto-generated, and include publication-ready figures, interactive plots, and robust variant and gene browsers.
For a more detailed introduction to Whole Genome Sequencing Analysis why not check out our Analysis Tutorial.
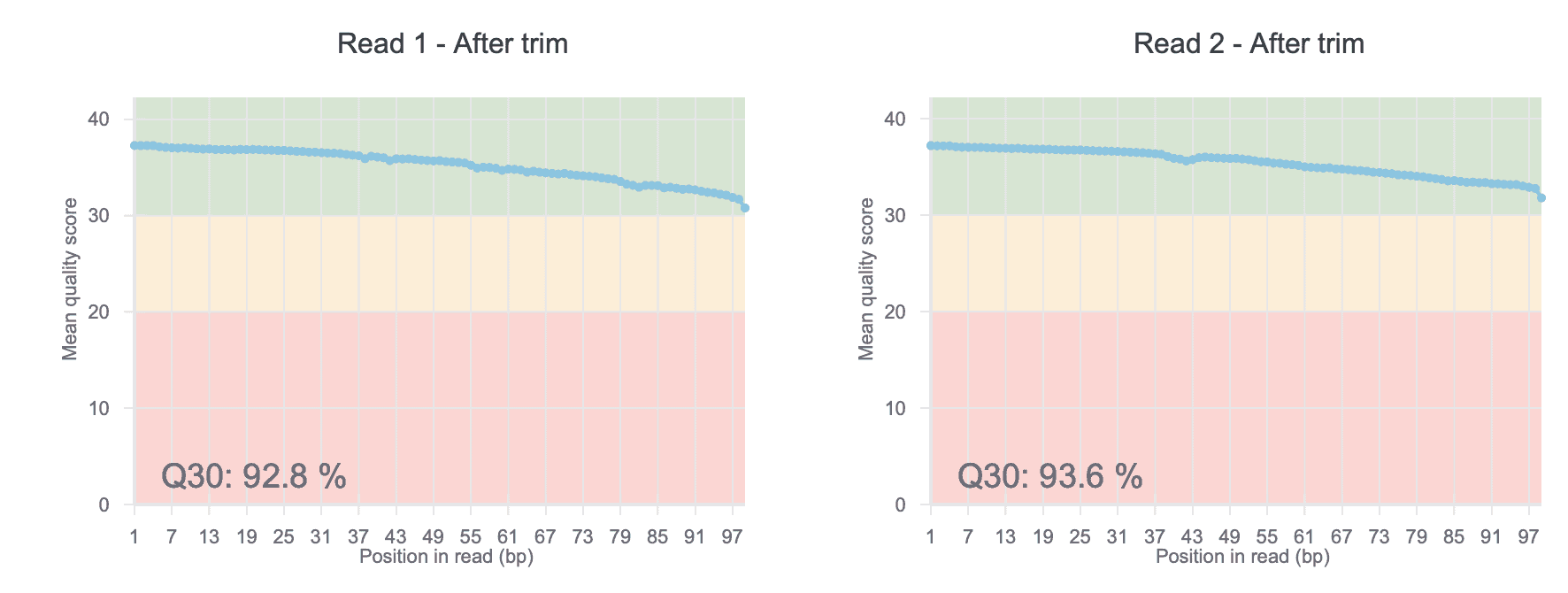
QC Plots
The quality of your DNA sequence analysis depends on the quality of your input. With each DNA-Seq report, Basepair provides useful QC plots that assign quality ratings to your data. You’ll quickly be able to spot any contamination and have an overview of base distribution.
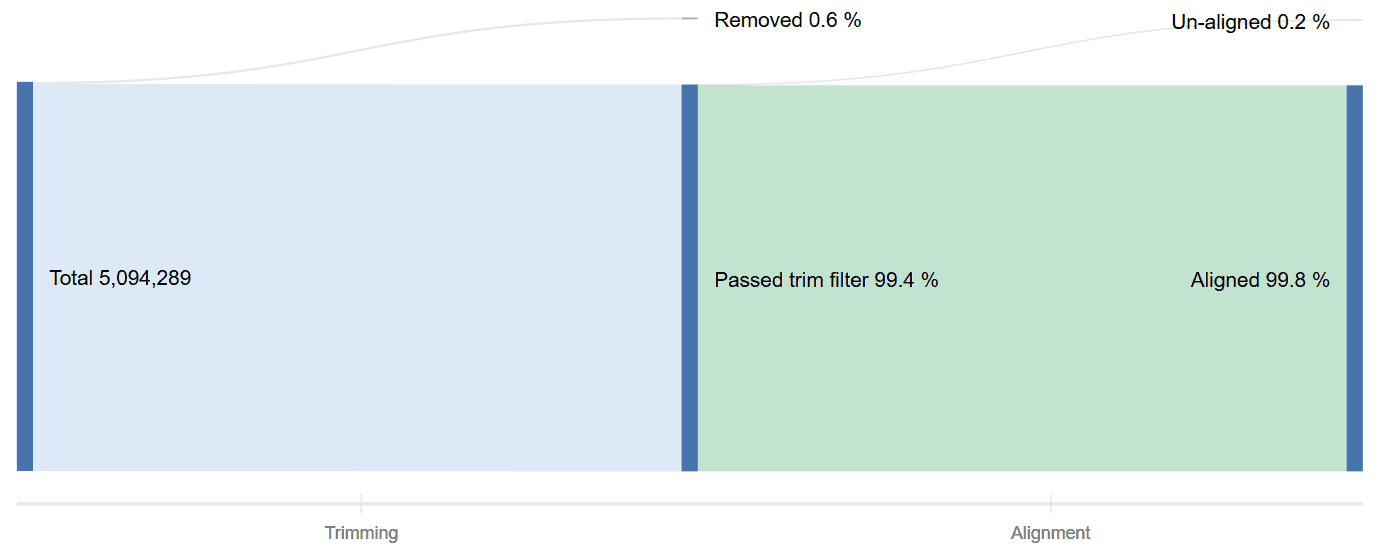
Read Counts
What portions of your DNA-Seq data was trimmed? Which reads were usable? The read count graph provides fast answers for any of your data quality questions.
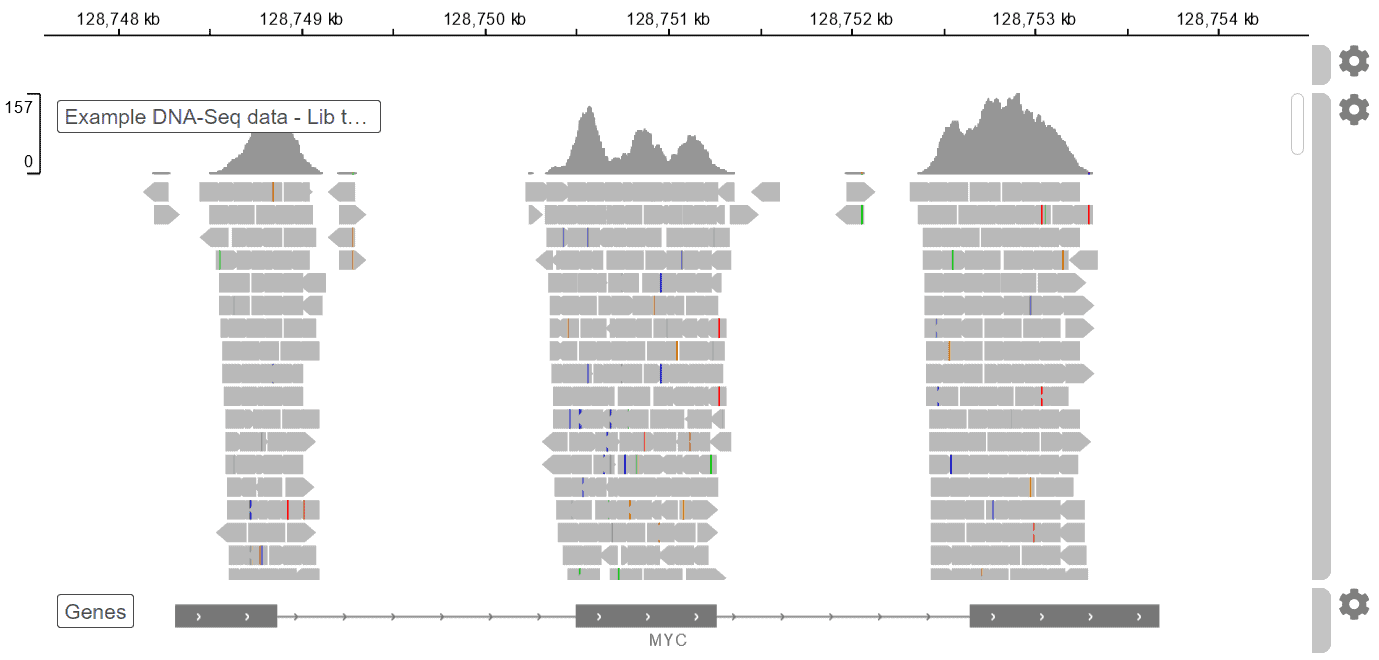
Genome Browser
We made visualizing genome coverage for various types of NGS data simple, interactive, and intuitive. All the technical tasks happen under the hood. Now you can explore all your raw DNA-Seq data, without the need for file conversions or downloading additional software.
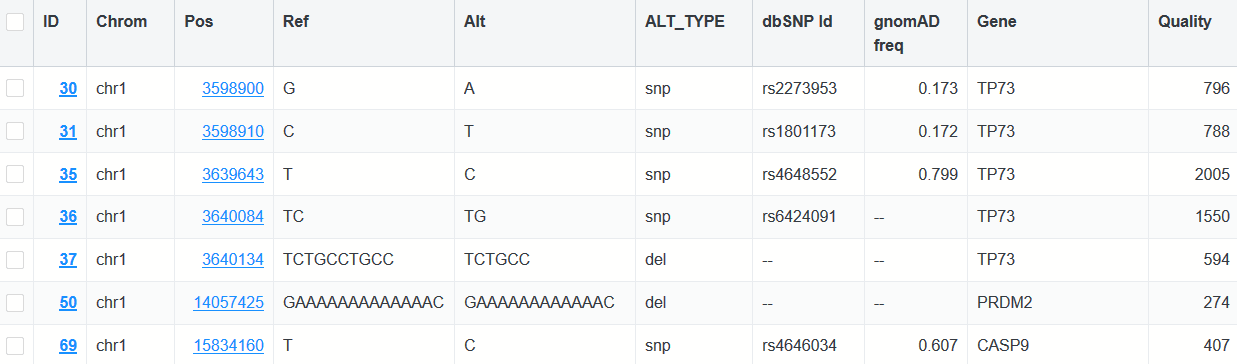
Variant Browser
Have a .vcf file and not sure what to do with it? The Variant Browser for whole genome/exome sequence data is a full-featured variant explorer with a collection of flexible parameters and filtering options, allowing anyone to quickly look through .vcf data and find what they need.
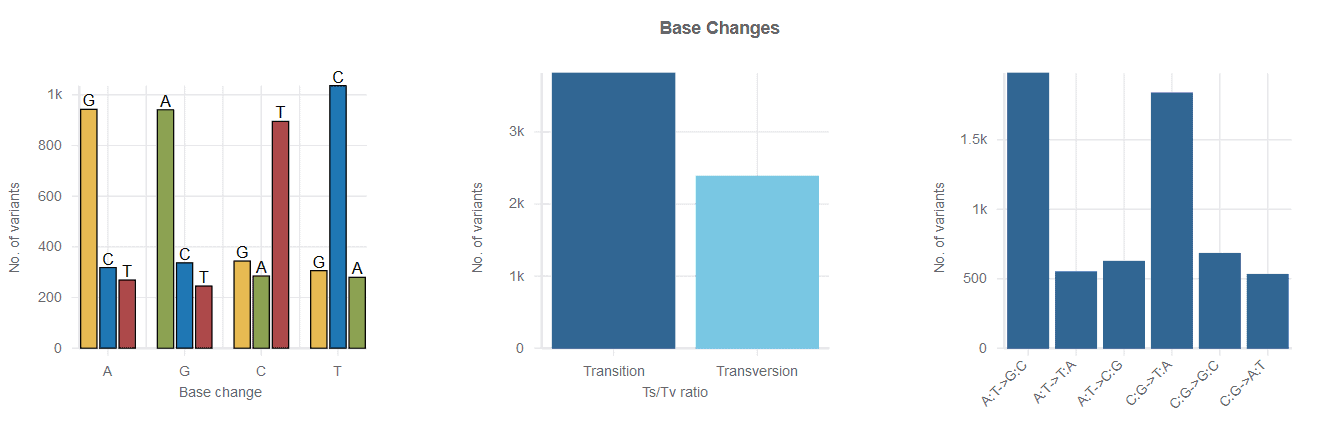
Variant Summary
In addition to the powerful Variant Browser, all variant workflows include a number of helpful metrics, such as gene annotations, base change counts, number of variant types, and much more.
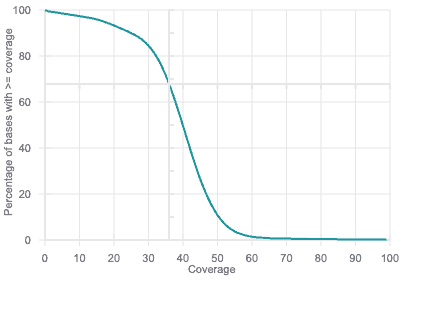
Coverage Plot
Basepair includes a coverage plot for your whole genome/exome data, pinpointing how well targeted regions are covered by data.
Set up your whole genome/exome analysis in minutes
Want to try these features for yourself? With our free 14-day trial, you can upload your own DNA-Seq data and choose among a number of battle-tested workflows, such as QC, alignment, variant annotation and variant calling, coverage, structural variants, and copy number. See why some of the world’s top institutions are using Basepair to save thousands of hours (and dollars) for their NGS data analysis needs.
Basepair Work is in Dozens of Peer-Reviewed Journals
"Fast, excellent and reasonably priced...you CAN get all three!! Thank you to the folks at Basepair for helping us deal with some difficult RNA Seq data."
"I really like how easy the website is to use. And how quickly the results are generated, including figures. I would have never thought about doing a new analysis like I just did."
"Support answers come fast and are always precise!"






